The Profiler module helps you to identify and quantify compounds in a sample spectrum, as well as apply spectral analysis techniques like spectral binning (PRO Only). You can fit any available compound to the current spectrum, and narrow down the compounds you're interested in by using compound sets and filters.
For a hands-on introduction to Profiler, see “Tutorial and Sample Files”.
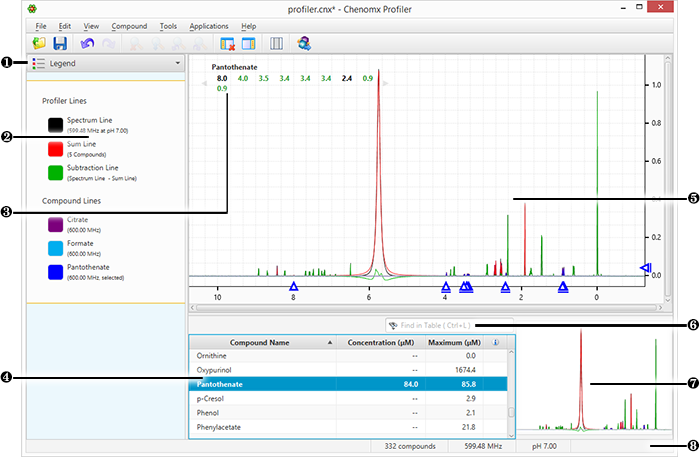
|
Sidebar View Dropdown Menu changes the currently visible Sidebar view. |
|
|
Legend customizes the lines appearing in the Spectrum View. |
|
|
Cluster Navigator navigates among the clusters of the currently selected compound. Enable or disable Cluster Navigator using the View menu. |
|
|
Compound Table displays compounds, their calculated concentrations, and their profiling status. Access compound signatures in the Spectrum Graph by selecting them. As well, you can view additional compound information in the Reference Panel by selecting a compound in the Compound Table and switching the sidebar view to the Reference Card Panel. You can change the default concentration units displayed in the Compound Table using the View menu. |
|
|
Spectrum Graph displays the spectrum as well as the sum line and subtraction line. Modify the display by focusing in on a particular area or display and modify compound signatures. |
|
|
Quick Search filers the Compound Table to locate particular compounds or groups of compounds. |
|
|
Spectrum Thumbnail displays a thumbnail image of the current spectrum. You can also use the thumbnail to navigate the Spectrum Graph at various zoom settings. |
|
|
Status Bar displays the current cursor position in the units specified by the View > X Scale and View > Y Scale settings. Also displays information about recent user actions. |
Compound Signatures with a peak-based fit style, such as those found in the Chenomx Reference Library, are fit to your spectrum by adjusting the compound height and making adjustments (sometimes called 'transforms') to the locations of their clusters. The result is a 'fit', or 'profiled' spectrum.
Peak-based compound signatures profile a spectrum by measuring compound concentrations by the height of each compound signature within the peak heights of your spectrum. When the height of a compound signature is adjusted, the corresponding heights of all peaks and clusters to that compound are modified.
The frequencies (i.e. positions) of the clusters in a compound signature may need to be adjusted during fitting due to slight mismatches in solution pH, ion concentrations, and other matrix effects. These potential errors are captured by a region around each cluster known as its 'transform window'. Profiler displays the transform window for each cluster as two triangles on the spectrum baseline when the cluster is selected. If the pH value for your spectrum has been calibrated correctly, you should be able to move each cluster to obtain a good fit without needing to move outside of the limits of the cluster's transform window.

