Version 10 of Chenomx NMR Suite introduces a new sophisticated, flexible and easy to use tool for automatic profiling of processed spectra.
Typically, when automatically fitting a spectrum, hundreds of variables (cluster positions and compounds concentrations) require optimization. Because compound concentrations are derived from existing peak cluster locations, and because peak clusters are allowed to be positioned within certain ranges (“transform windows”), several rounds of optimization are necessary to achieve a local or global minimum, i.e. a solution that reconstructs the experimental spectrum as best as possible.
Moreover, different mathematical solutions may recreate the experimental spectrum with similar degrees of fitness/fidelity. For this reason, the software can explore those solutions by repeating the fitting process for each individual input file as many times as the user chooses, and output a solution for each pass.
It is important to keep in mind that automatic profiling is purely a mathematical process that simply relies on a provided compound library. It does have not have the prior chemical knowledge of a human operator. COMPLETE Autofit should not be used blindly and fit results should be reviewed manually.
Steps to set up COMPLETE AUTOFIT
-
Open your processed CNX file within the Profiler Module.
-
Go to Tools -> Batch Operations -> "COMPLETE Autofit"
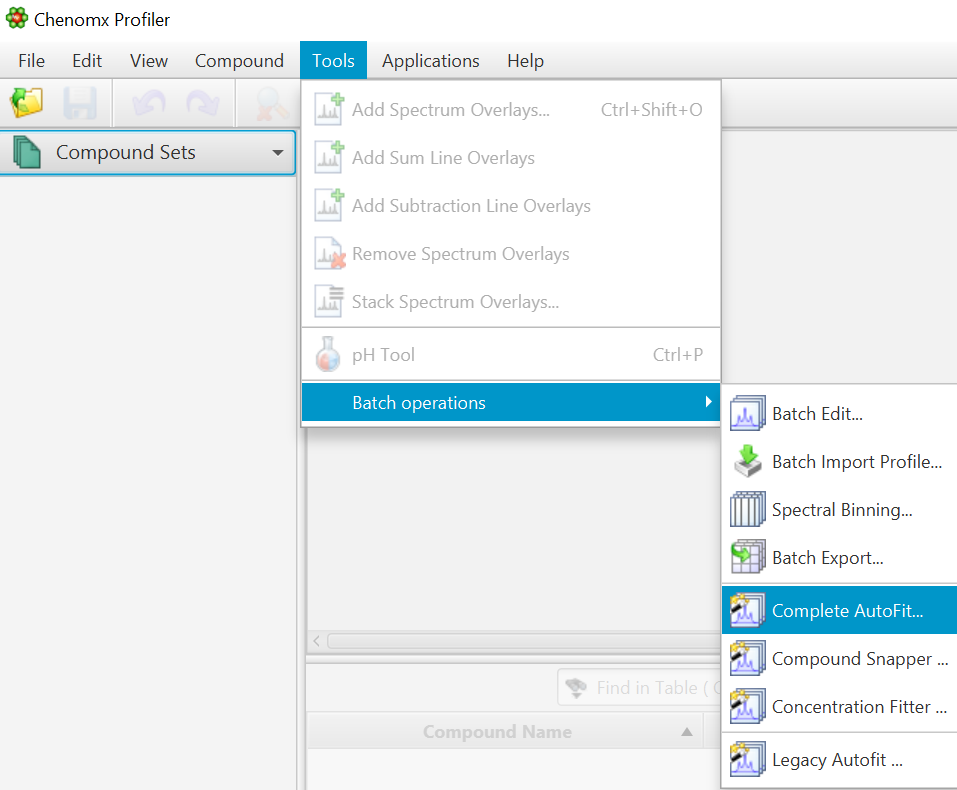
-
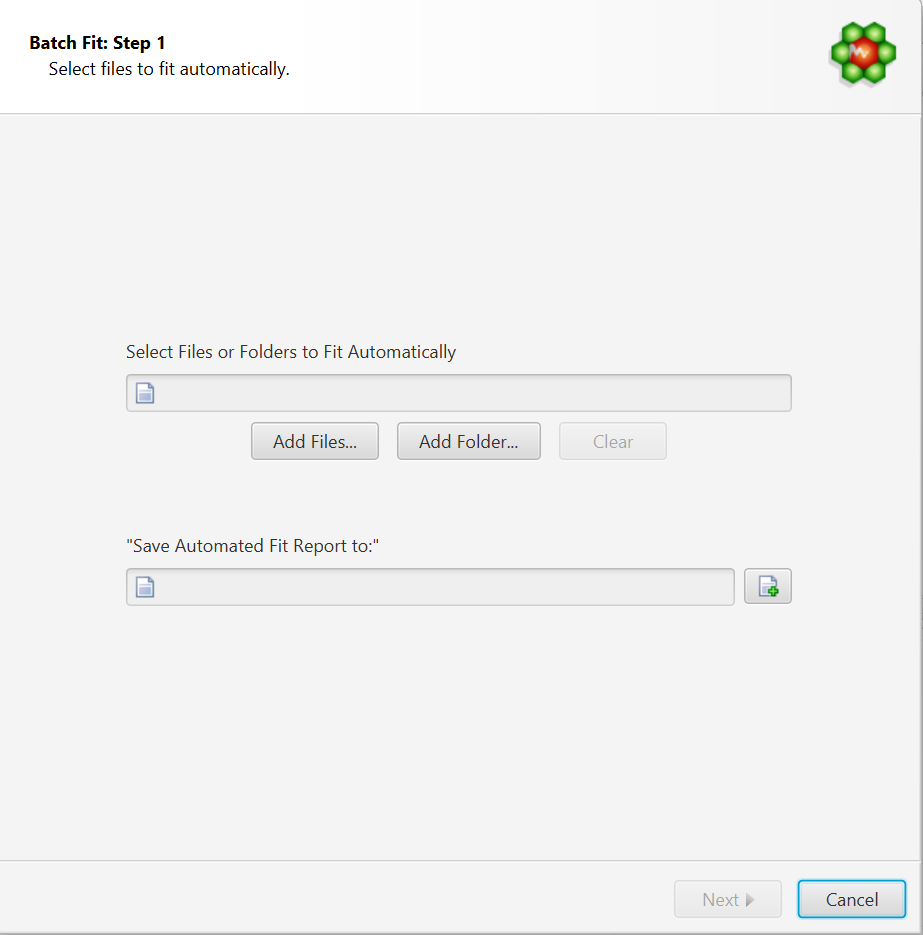
Select Files to Fit Automatically
-
Select Files or Folders to Fit Automatically: COMPLETE Autofit will do a complete profile of one, or several CNX files. Select your unprofiled CNX files here.
-
Save Automated Fit Report to: A fit report in the form of a tabulated separated value (tsv) file will be generated, with each input file and the sum of residuals associated with each fit attempt. Specify the destination and name of the report file here.
-
-
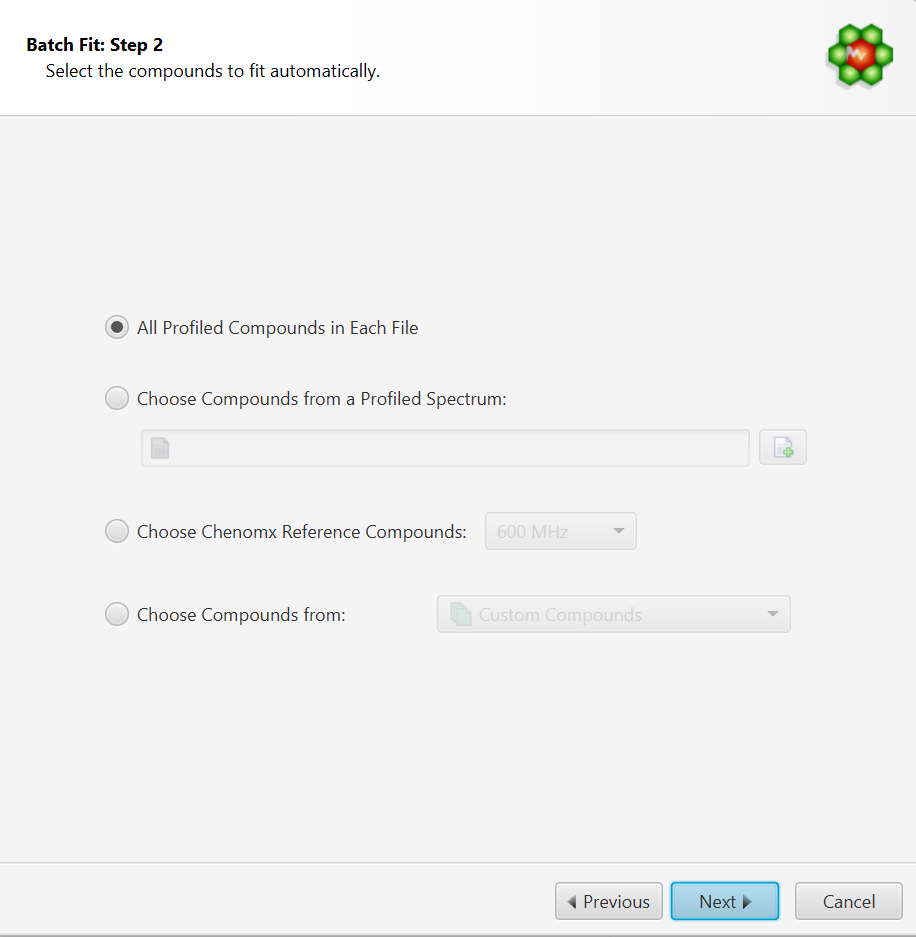
Select the Compounds to Fit Automatically. You can either:
-
Choose to fit all the compounds already profiled in each file. In this case, compounds must be already present in their profile and all of the compounds will be included in the fit operation.
-
Choose compounds from and already profiled spectrum. Note that it is possible to select only a subset of those in the interface’s next step.
-
Choose compounds from a Chenomx library
-
Choose compounds from a customized compound list defined in Library Manager.
IMPORTANT NOTE: DO NOT ATTEMPT TO FIT THE ENTIRE CHENOMX LIBRARY. THIS WILL RESULT IN OVERFITTING AND AN EXTREMELY LONG PROCESS. TARGET THE COMPOUNDS THAT SHOULD BE VISIBLE AND PRESENT FOR THE TYPE OF SAMPLES YOU HAVE.
-
-
Compound Selection Refinement
-
• Depending on your selection in the previous step, you may be presented with an option to further refine your compound selection. You can move compounds across list boxes to specify whether or not you want to include them in the fit.
-
• The checkbox option “Skip compounds that are already fit” is self-explanatory. It allows you to ignore compounds that are already fit. Their contribution will be calculated and taken in consideration to fit the reminder of the compounds, but their current state (cluster positions, concentrations) will not be modified.
-
-
Concentration Fitting Setup
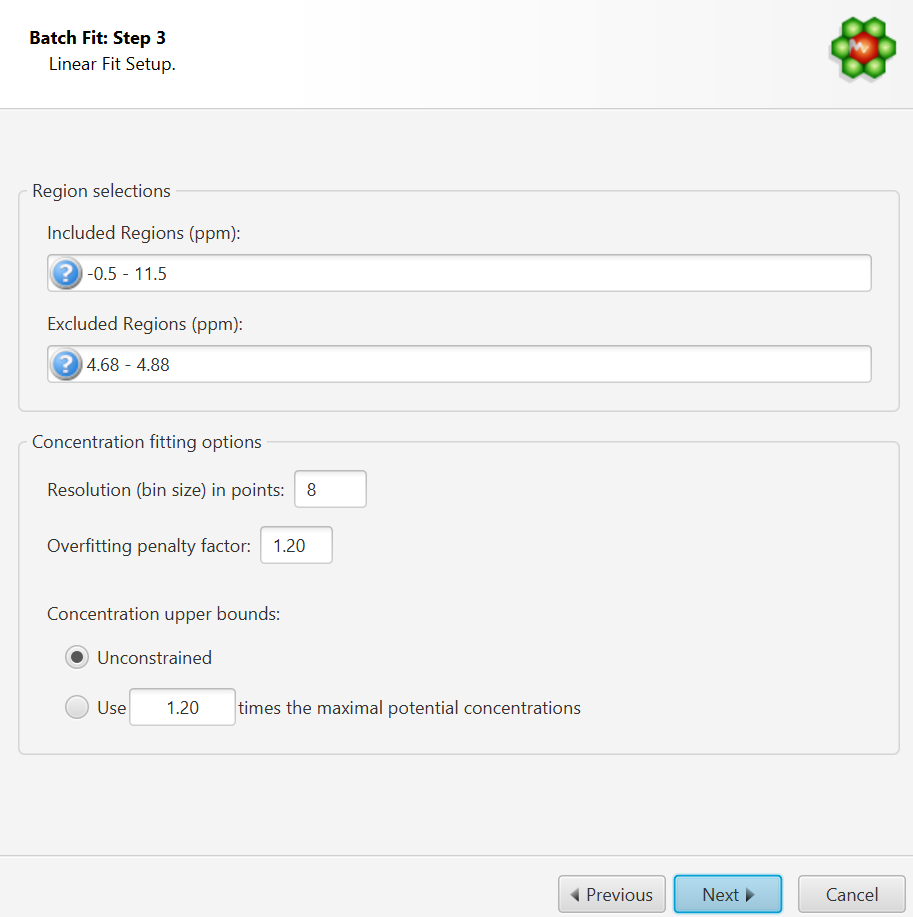
This interface is composed of two main sections: Regions Selection, and Concentration Fitting options. The role of each section is described below. The application Preferences panel can be used to control and set all the default values appearing in the interface.
-
Region Selection: Comma-separated list of spectral regions (in ppm units) to be included or excluded in the fit. Each component of the lists must include a lower and upper bound in ppm, separated by a dash. For instance, “0.8-2.3, 3.0-4.2” is a valid list.
-
The Included Regions input field displays the limits of the currently selected spectral region (as selected with the Select Region Tool). If no region is currently selected, the Included Regions input field defaults to the entire spectrum.
-
The Excluded Regions input field will automatically include the region associated with the water peak. It is recommended that any peaks around the water region be excluded due to attenuation caused by water saturation. Similarly, the region associated with urea (5.4-6.0 ppm) should also be excluded if urea is present.
Excluded regions have prevalence over any included regions, i.e. any spectral point falling within any of the listed excluded regions will automatically be ignored during the fit.
-
-
Concentration Fitting Options: This group of options control the behavior of the optimization engine.
-
Resolution(bin) size:
The spectrum can be fit using either its full representation, or a binned representation. This parameter controls the number of spectral points per bin. Set this parameter to 1 for full resolution. However, it is highly recommended to use a binned representation if the peak cluster positions have not been optimized. Fitting at full resolution will require more memory and CPU time.
-
Overfitting Penalty Factor:
The amount of overfitting can be limited by increasing the weights of the spectral points where overfitting occurs. This will effectively increase the importance of the spectral points where this occurs relative to the other ones, adding a penalty to pay for predicting points over the spectrum line. For a given spectral point, the square of the difference between the experimentally observed spectrum intensity and the predicted/calculated one will be multiplied by the factor specified in this field if the difference is negative. The weights are adjusted iteratively and up to 5 cycles of optimizations are performed if the specified penalty factor is >1.
-
Concentration Upper Bounds and Penalty Factor:
Non-negative constraints are used during the optimization process so that concentration values remain ≥ 0. Two different settings are offered to control their upper bounds. They can be set to be unlimited via the Unconstrained option, or limited according to their predetermined maximal potential concentration. The maximal potential concentration of a given compound is the highest concentration for which the compound signature would not exceed the experimental spectrum.
Because of possible discrepancies between the compound library signatures and the experimental spectrum, mainly due to pH conditions, matrix effects, etc, the potential concentrations determined by Profiler may not be optimal and should only be regarded as guidelines. To give potential concentrations more wiggle room, potential concentrations can be pre-multiplied by a specified constant. The derived set of scaled potential concentration values will then be used as upper bounds.
-
NOTE: It very important to note that the baseline is not modeled from the residuals at any stage. It is therefore crucial that the baseline be properly corrected prior to concentration fitting.
-
-
-
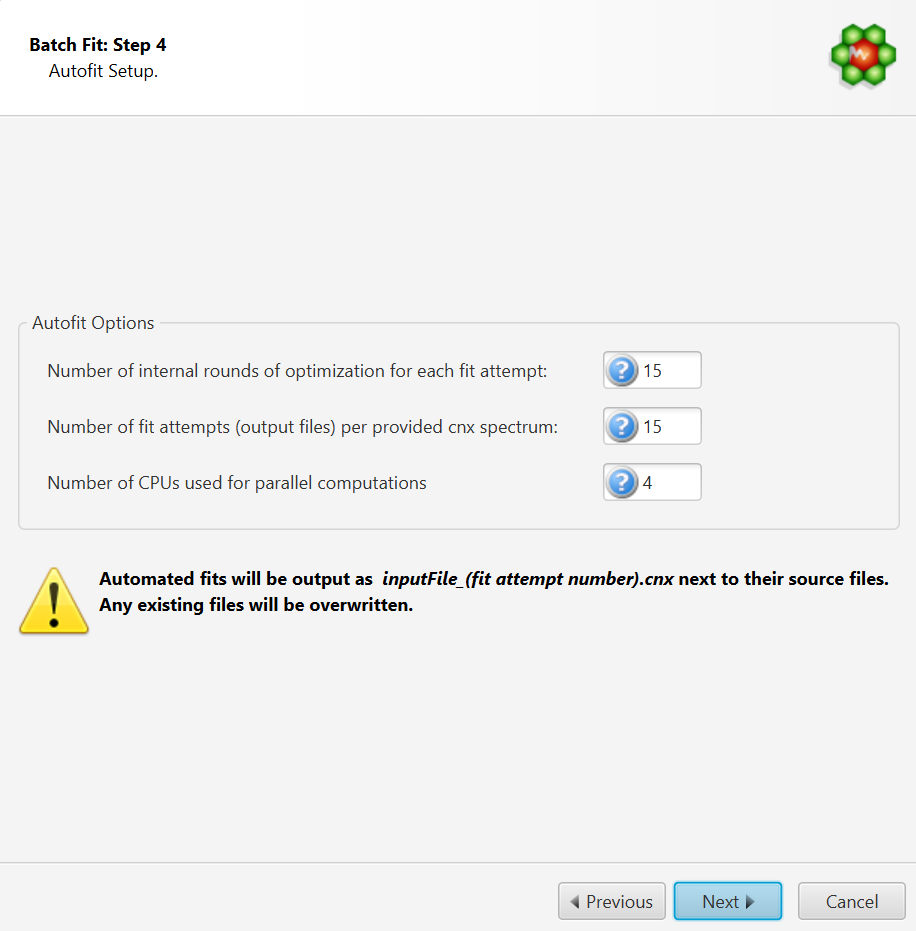
Autofit Setup
This panel controls the overall behaviour of automated fitting:
-
The number of internal iterative rounds is related to the number of times, for each fit attempt of a given spectrum, the algorithm will position clusters and adjust concentrations. It is directly equivalent to the situation encountered with manual fitting, where an operator usually has to do several rounds of going through the list of compounds to be fit and adjust their peak cluster positions and height (concentration).
-
The number of fit attempts control how many output files will be generated for each input spectrum cnx file. By analogy, it is related to how many people would be trying to fit the spectrum if there were a team of operators. Each one could potentially come up with a different solution that produces more or less the same fit quality in terms of sum of squares of the residual line.
-
The number of CPUs control how many CPUs can be used for parallel computations. The number presented in the interface takes into account the maximum number of CPUs allowed and defined by the software license, and the number of CPUs that may already be used by another COMPLETE Autofit job.
-
-
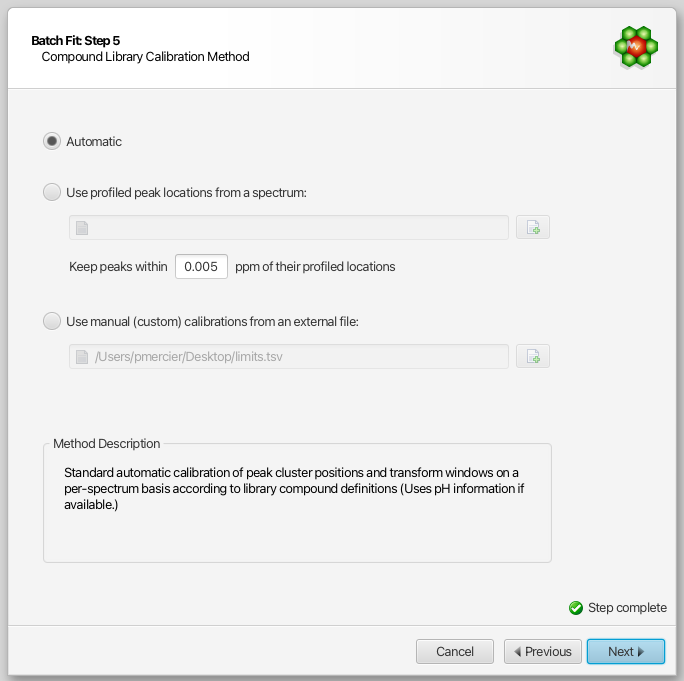
Compound Library Calibration Method
The compound library is automatically calibrated on a per-spectrum basis. The starting peak cluster positions, as well as their range of allowed values (transform windows), are determined based on pH information in each spectrum. Optionally, you can overwrite the positional restraints with your own set of values.
This can be particularly useful in situations where peak multiplets appear outside of their calibrated transform windows, due to matrix effects or other causes. Although Profiler allows for manually positionning multiplets outside of their transform windows, the AUTOFIT procedure strictly enforces them.
A mechanism was put in place to overwrite (post-calibrate) the automatically determined values. You can force the automated procedure to position multiplets around their profiled locations in an external spectrum, within a give range (in ppm units). This method is compound-based, i.e. all clusters of a given compound will be restrained.
Alternatively, you can opt for a more advanced and flexible cluster-based approach, via an external tab-separated file prepared within Profiler. Refer to the section “Custom calibration of peak cluster centers ranges for automated procedures” for the preparation of such a file.
You can further refine your compound selection in the following next step.
-
Compound Selection for Custom Compound Library Calibration
If you have choosen to post-calibrate (overwrite) the automatic compound calibration with your own set of values, you can further refine which compounds it will apply to from the all of the compounds available from your selected source in the previous step:
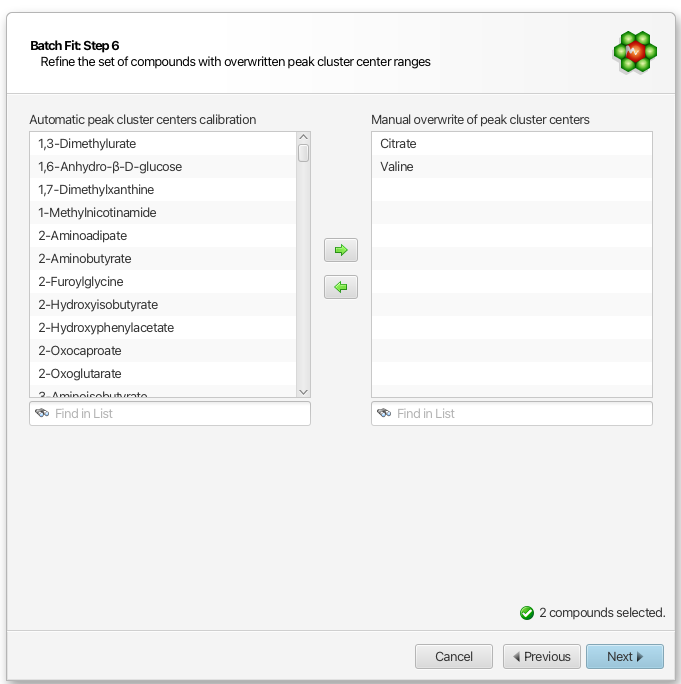
At this step, you should exclude compounds known to be particularly very sensitive to pH (exhibit large chemical shift variations with pH). For instance, it is highly advisable to exclude any of the histidine-based compounds if you are operating around pH 7.0, and simply let the automated procedure proceed with the default calibration for those.
Important: If you choose a profiled cnx file as your source of calibration, all the clusters of the selected compounds in this step will be considered, whereas if you choose an external tab-seperated value file, specific clusters can be included or omitted.
-
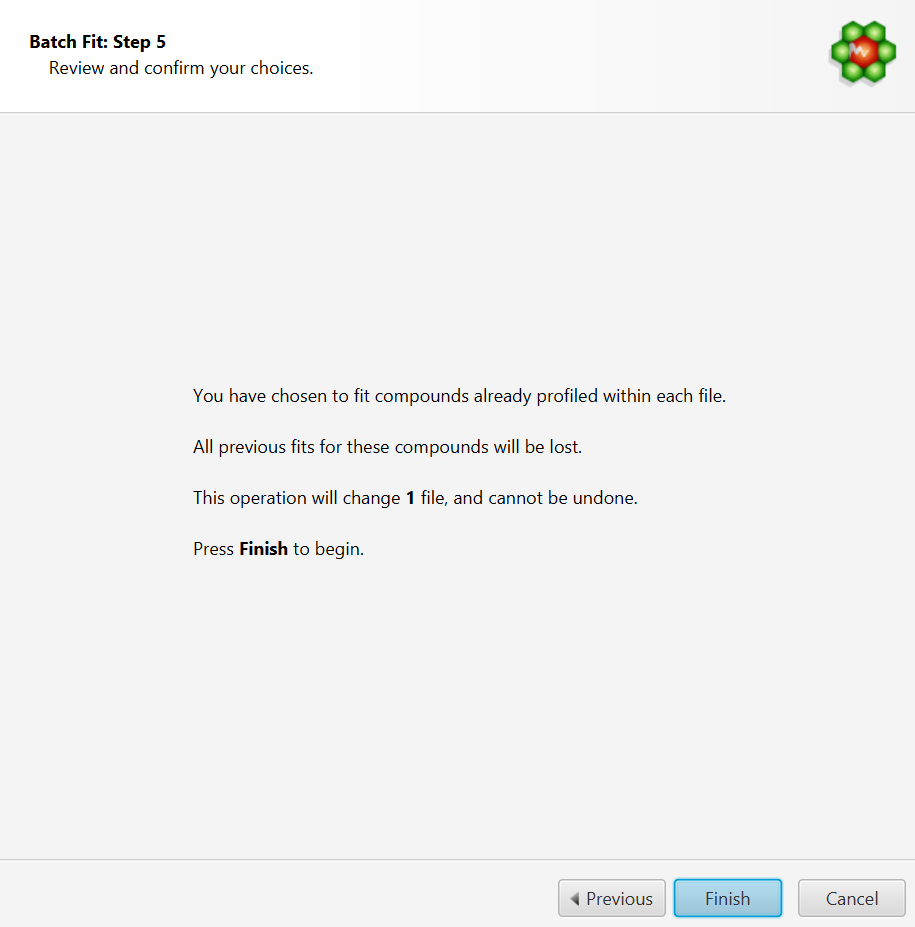
Review and Finish
-
COMPLETE Autofit will generate multiple profiled CNX files for each input spectrum file that will be saved in the same location as the input source. It will not however modify the original source files themselves.
-
A fit report will be produced, showing the residuals chi-square value for each of the produced cnx file. For a given input file, the output solution with the lowest chi square (lowest fit error) is automatically saved with the label “_best” appended at the end of its filename.
-
A text file (.txt) will also be generated for each exported CNX file with the chi square value achieved for each internal round of iteration. Inspection of those text files can reveal whether or not the number of internal rounds that were specified is enough to achieve a minimum. If the best solution (lowest chi square) is generally obtained only at the last internal iterations, it is a sign that maybe a higher number of iterations is needed.
-
It is up to the user to choose the output solution that produced the best fit, if the decision parameter is other than the sum of residuals. For statistical analysis (PCA, PLS-DA, etc.), it may be more relevant to work with concentrations averaged over the series of exported fits rather than single values.
-
-
Not all spectra are suitable for automated fitting. Unfiltered spectra showing presence of larger molecules though humps will likely fail. The software does not model the baseline on the fly.
-
The raw fid must be correctly processed and the baseline must be adjusted properly. The software does not model the baseline on the fly.
-
Set the pH for each sample, either in Processor or Profiler. pH information is used to calculate the boundaries for each peak cluster. The software will not fit peak clusters outside of their calculated transform windows.
-
If spectral regions contain intense unknown peaks, exclude those regions from the fit in the provided interface.
-
The time required to fit a spectrum is directly dependant on the number of spectral points, and the total numbers of peak clusters and compounds. Avoid under and over fitting, i.e. work with compound subsets that represent what you should expect to find in the type of samples you are analysing.
Version 10 also reintroduces a form of automated fitting found in version 8.6 and earlier of the software, as “Legacy Autofit”. Legacy Autofit may be preferable to older users that want to maintain controls for a project initiated using older versions of Chenomx NMR Suite. For users starting a project using Chenomx NMR Suite Versions 9 or later, consider using more advanced Autofit techniques such as Compound Snapper / Fit Automatically or the COMPLETE Autofit Tool.
The difference between Legacy Autofit and COMPLETE Autofit is explained below.
-
Legacy Autofit first takes the raw spectrum and subtracts the contribution of all previously fit compounds that are not selected for the autofit.
-
Starting from the spectrum obtained in step #1, it scores the compounds selected for autofit to establish the order in which they will be fit.
-
Starting from the best potential candidate, it tries to simultaneously establish the position of the clusters for that metabolite and the maximum intensity (and therefore concentration) of that compound on the spectrum. (If it fails to establish where some clusters are located, they are left at their default/untransformed locations.)
-
The contribution of the potential candidate is then subtracted from the spectrum to yield another spectrum which serves as starting point for the processing of the second candidate compound, using the same procedure.
-
This cycle is repeated until the list of selected compounds for autofitting has been exhausted.
-
One can clearly see that the risk of errors increases at every step, and that eventually not all compounds will succeed in being autofitted. Moreover, as individual compounds are added at their maximal concentrations, the final result does not reflect a global optimization of all compounds, but reflects a successive series of individual optimization. (In order words, after adding compound #x, the algorithm does not reevaluate compound #x-1, #x-2, #x-3, etc.)
The main difference between COMPLETE Autofit and the Legacy Autofit approach relies on the fact that the new algorithm is based on a true “global” optimization process. The compound concentrations are obtained altogether (globally) using penalized least squares, rather than through successive fit rounds of individual compounds. The algorithm used for positioning peak clusters has also been greatly improved. Be aware though that there is a computational speed cost to those improvements.

