NMR Metabolomics
quantified
Chenomx specializes in mixture analysis for applications in life sciences such as metabolomics, food/nutrition, and cell culture research.
Free Evaluation Version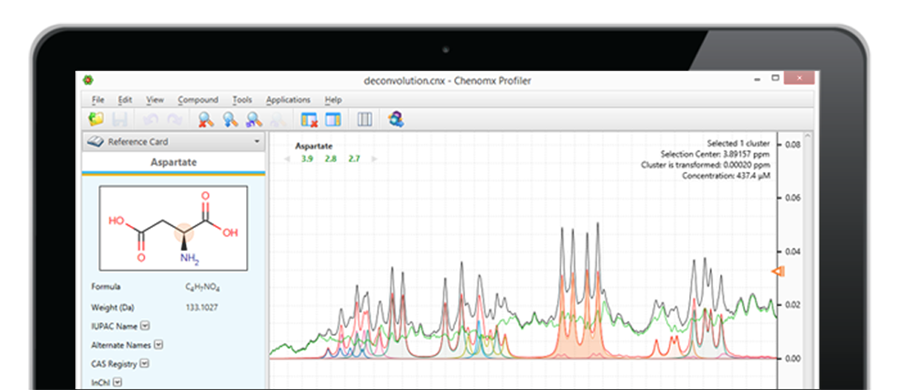

Why Chenomx?
Used by Metabolomics researchers world-wide, Chenomx NMR Analysis Software works with comprehensive Spectral Reference Libraries to both identify and measure concentrations of compounds visible in the NMR spectra, all in one integrated workflow. This patented software offers best-in-class concentration measurement, identification and advanced de-convolution of spectra.
Chenomx NMR Analysis Services are available, providing cost effective and efficient solutions for mixture analysis of client samples.
De-Convolution Method Exposes and Quantifies Overlapping Signals
Proprietary Reference Libraries Supporting 400MHz through 800MHz NMR Instruments
Concurrent Identification and Concentration Measurement
"Chenomx has a seamlessly designed user interface that is quick to learn and easy to use. It has made NMR metabolomics vastly more accessible to researchers. Our undergraduate and graduate students operate Chenomx independently, identifying and quantifying metabolites for their research publications."
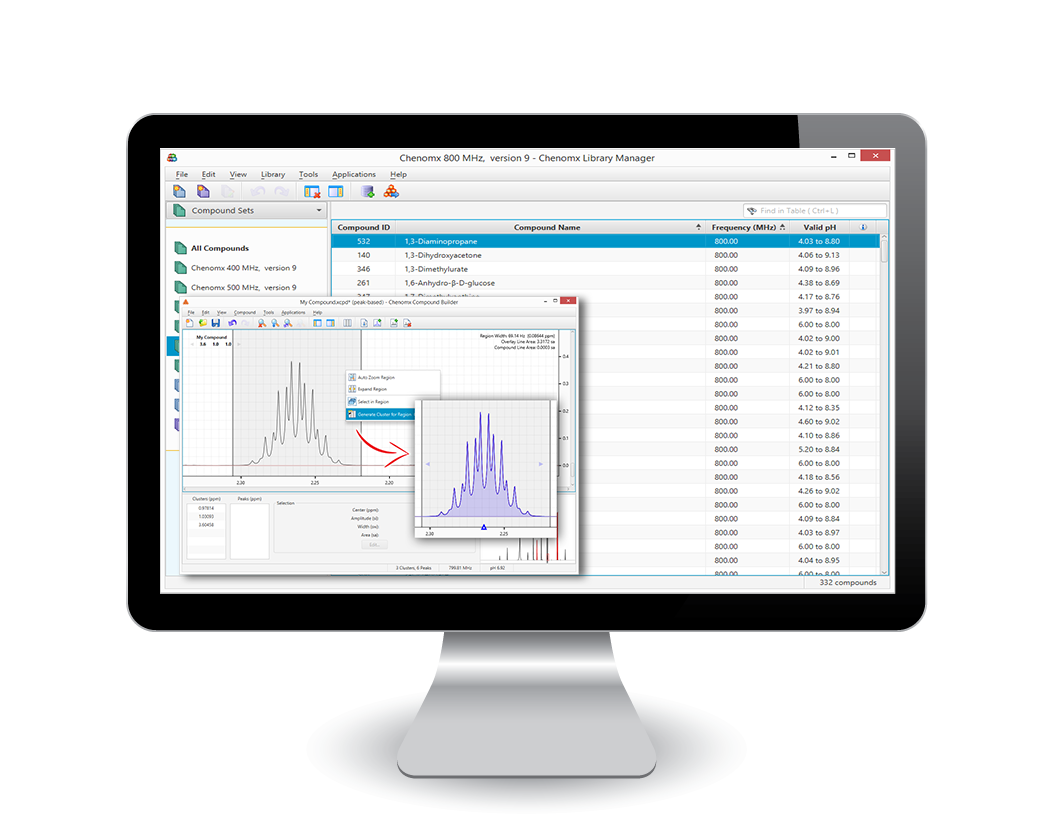
The Chenomx Library
The Chenomx Spectral Reference Libraries are developed from thousands of reference spectra obtained through NMR analysis and used for identification and comparison purposes. The Reference Libraries are available from pH 4 through 9 and NMR field strengths from 400 MHz through 800 MHz.
Additional Reference Libraries have been created from the Human Metabolome Database (HMDB.ca) and are also available for download (Chenomx cannot certify these entries, but they can be used for ID purposes).
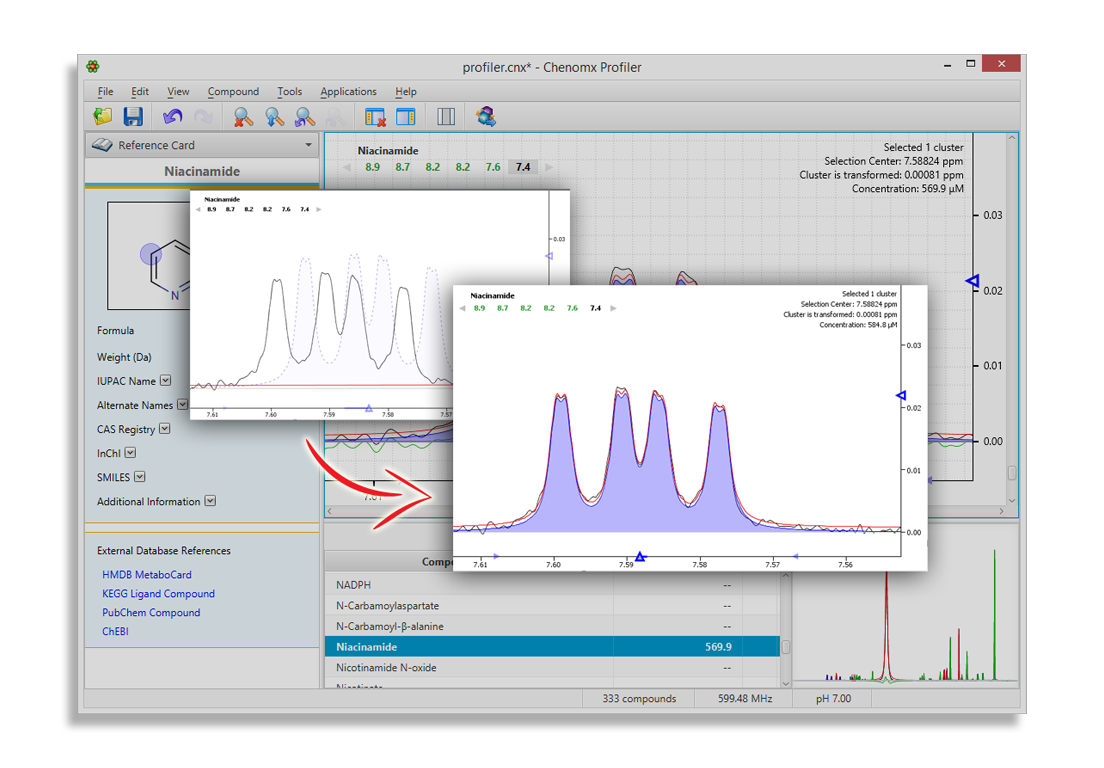
‘Fitting’ the Signatures to the Spectrum
Chenomx NMR Analysis Software functions by 'fitting' the Spectral Reference Library to the appropriate signals within the experimental spectrum. The software automatically adjusts the Reference Library to reflect the sample and acquisition conditions (pH, NMR field strength, spectra line width) and allows the used to adjust peak intensity and location for fine-tuned concentration values.
After the experimental signal has been fit, the spectrum's residual line is exposed. The profiling software will compare more compounds from the Spectral Reference Library to isolate and identify the remaining signals.
"Chenomx is the only commercial software that I use in my metabolomics pipeline. It is easy to use and exceptional at absolute quantification. With its well curated library of spectra one can deconvolute complex and variable samples accurately and with high confidence. It also allows the identification of less abundant compounds in convoluted or highly overlapped areas of the spectrum."
Applications
Cell Culture
Metabolite analysis has proven to be an invaluable tool in observing and understanding the metabolic phenotype of cells in culture. In large bioreactor conditions, there are many challenges such as optimizing growth media, growth feeding strategies, and other engineering parameters that are vital not only for the growth of the cells but for the production of your products. With Chenomx's Profiling technology, getting reliable and quantitative data is key to overcoming many of these challenges.

Food and Nutrition
From essential amino acids to various sugars and other important components of food such as antioxidants, quantitative measurements of nutritional content of food is key to understanding the differences between food products. Understanding diet related diseases or growth factors, and their correlation to food intake is vital in our strive for better health. Earlier nutritional intervention can possibly prevent the onset of diseases that may require pharmaceutical aid in the future.

Drug Metabolism
As drug development costs skyrocket, it is ever so vital to fully understand your drug targets. Metabolomics can be a critical tool in drug discovery. From early detection of toxicity to mechanisms of action at a cellular level, positively identifying and quantifying metabolites in relevant biofluids can bridge the gap of understanding of your compounds. From preclinical to clinical stages of drug development pipeline, extra quantitative metabolite data will add real value to your drug submissions. Chenomx has years of experience with serum, plasma, urine, CSF, saliva, tissues, and cell based models.
Biomarker Discovery
Discovering new biomarkers has been a goal for clinical researchers to understand disease mechanisms and aid in the treatment of these diseases. Not only does NMR offer an analytical method to encompass a wide range of molecule types, but coupled with Chenomx technology, this technique becomes even more valuable as identification and quantification of the components in the samples are discovered. Chenomx has years of experience designing experiments, building diagnostic models, and validating them with clinical trial data.